BITOLA - Biomedical Discovery Support System
By Dimitar Hristovski & Borut Peterlin
Purpose
BITOLA is an interactive literature-based biomedical discovery support system. The purpose of the system is to help the biomedical researchers make new discoveries by discovering potentially new relations between biomedical concepts. The set of concepts currently contains MeSH (Medical Subject Heading), which is used to index Medline, and human genes from HUGO. The potentially new relations are discovered by mining the Medline database.
To make the system more suitable for disease candidate-gene discovery and to decrease the number of candidate relations, we integrated background knowledge about the chromosomal location of the starting disease as well as the chromosomal location of the candidate genes from resources such as Entrez Gene, HUGO and OMIM. The BITOLA system can also be used as an alternative way of searching the Medline database.
To understand the details of the system and see some examples of its use, you are strongly recommended to read these papers.
The two BITOLA tools
The system is available in two versions: "closed discovery" and "open discovery".
Closed discovery allows the input of two concepts (Example 1: a disorder and a gene. Example 2: a drug and a side effect) and generates potential explanations of the relationship between two entities. It does this by searching published literature to finds intermediate links.
Open discovery allows the input of a single concept, then categories for first-order relatives of that concept, then categories for relatives of those first order concepts. Thus it can link from a disease to related drugs, then to genes related to those drugs and then test if those genes have been mentioned/tested in association with the disease. If the answer is no, then the gene is potentially related yet untested in the literature. Thus the open discovery tool is a nominator of new genes, drugs or neuroscience correlates to be investigated with diseases, disorders, physiological responses or any other phenotype.
Please click on the pictures to access the following tools:
(NOTE: If you have not used BITOLA before, please read the tutorial below first)
Tool 1: Closed discovery system Tool 2: Open discovery system
Tutorial
Literature-based discovery (LBD) is about finding connections between otherwise disconnected areas of literature. The field began in 1983 with Don Swanson who came across two pieces of literature which were clearly related, but had not been linked. One paper described how fish oils thin the blood - another paper evidenced that Reynold's Disease was associated with thicker blood. However, it appeared as though no one but Swanson had read both papers and realised the clear implication that fish oils may ammeliorate Reynold's Disease. After investigation, this turned out to be the case. Swanson (and later, others) then made efforts to generate bioinformatics tools to uncover other similar "discoveries" latent in the literature, but as yet not explicitly mentioned or tested. This is the aim of modern literature-baesd discovery tools such as BITOLA.
Using the Closed Discovery System: a "gene explanation" example.
When you open the closed discovery tool, the screen will look something like this:
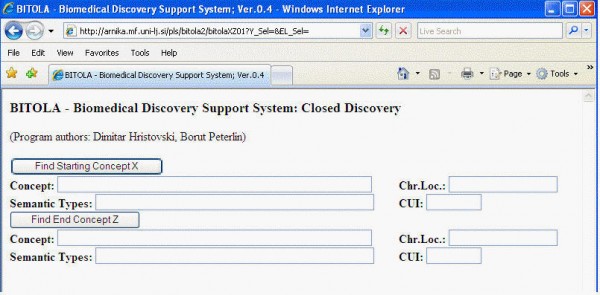
This is a short description of a typical use of the system. Let us say that you have found an association between clinical depression and the gene DRD4 and would like to know the neuroscience of why this might be:
- In the top concept box, type DRD4 and click "Find Starting Concept X".
- From the generated list, choose the gene DRD4. You will see that the chromosomal location is automatically filled in, as is the semantic type (gene or gene product)
- In the next box, type in "depressive" and click "Find End Concept Z".
- Now choose "Depressive Disorder" (official MeSH term) from the generated list. You will see the box Semantic Types automatically fills with "Mental or Behavioral Dysfunction". Also, a new box appears showing you various ways to limit the search for links between X and Z.
- For now, go to the Sematic Type pull-down list in the new box, and from the alphabetically-ordered list of terms, choose "organic chemical".
- Now click the button "Find Intermediate Ys"
-
A list of organic chemicals which have been linked to DRD4 in some papers, and Depressive Disorder in other papers, is generated. The screen now looks like this:
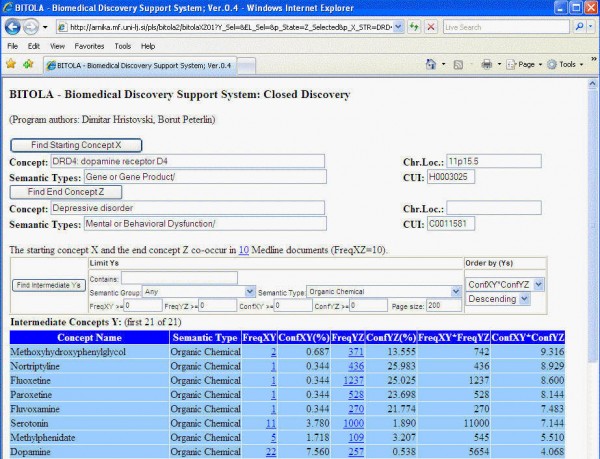
- With the top chemical returned (Methoxyhydroxyphenylglycol), click on the hyperlinked "2" in the FreqXY column; This takes you to the two papers in PubMed which were written about both your X concept (DRD4) and this Y term (Methoxyhydroxyphenylglycol)
- You can now explore other links, or re-run the query with other categories, exploring all domains and chemicals/neuroanatomy which have been linked to both DRD4 and Depressive Disorder.
Once you have tried using the Closed Discovery system, you will find the Open Discovery System to be very similar. The only difference is the structure. With closed discovery you nominate X and Z then search in Y (limiting categories, if desired). With open discovery, you nominate X, then search in Y (limiting categories, if desired), then search in Z (limiting categories, if desired).
Publications
-
Hristovski D, Friedman C, Rindflesch TC, Peterlin B.
Exploiting semantic relations for literature-based discovery.
AMIA Annu Symp Proc. 2006;:349-53. (PDF) -
Hristovski D, Peterlin B, Mitchell JA, Humphrey SM.
Using literature-based discovery to identify disease candidate genes.
Int J Med Inform. 2005 Mar;74(2-4):289-98. (PDF) -
Hristovski D, Peterlin B, Mitchell JA, Humphrey SM.
Improving literature based discovery support by genetic knowledge integration.
Stud Health Technol Inform 2003; 95 68-73. (PDF) -
Dimitar Hristovski, Borut Peterlin, Sašo Džeroski, Janez Stare.
Literature Based Discovery Support System and its Application to Disease Gene Identification.
Book chapter in print describing in detail an earlier version of the system. (PDF) -
Hristovski D, Stare J, Peterlin B, Dzeroski S.
Supporting discovery in medicine by association rule mining in Medline and UMLS. Medinfo. 2001;10(Pt 2):1344-8. (PDF)
Take me to the IBMI webtools homepage.